
Introduction
CRISPRcruncher takes a DNA sequence up to 99 base pairs in length, which encodes a peptide (up to 33 amino acids in length) and reports all possible restriction sites that could be created by changing one or more nucleotides while preserving the original peptide sequence. The tool takes advantage of the redundancy in the genetic code along with a built-in library of 643 commercially available enzymes. Note that commercially available enzymes may change with time and that certain enzymes may not be available in all countries.
The reports for creating a new sites always show the most parsimonious solution (i.e., the fewest possible substitutions). The report also indicates if the restriction site corresponds to the entered top strand, the bottom complementary strand, or to both, as in the case of palindromic sequences (e.g., GAATTC). CRISPRcruncher will also report restriction sites that are already present in the original sequence, but does not provide suggestions for altering such sites, as this is easy to do manually.
CRISPRcruncher should work on all browsers, although this may depend on the version you are using; certain versions of Safari may be problematic. CRISPRcruncher has worked on all versions of FireFox and Chrome that we have tested.
Directions
- Enter some personal information, then enter a nucleotide sequence from 15 to 99 base pairs in length.
- The sequence length must be a multiple of 3.
- To provide a useful output, the entered sequence must start out in the correct frame. For example, if the first amino acid is methionine, then the first three nucleotides of the sequence should be ATG. Note that stop codons are treated identically to amino-acid codons by the tool.
- Choose a minimum length recognition sequence for the reported restriction-enzyme sites. Choosing 4 (i.e., 4-cutters and larger) will give you the most options. Choosing 5 or more will provide fewer options, but those it provides may be the more useful ones. Note that the sequence CCGG and CCNNNNGG are both considered to be 4-cutters. Additionally, non-specific nucleotide designations, such as R and Y, are counted the same as G, A, T, C. For example, AARYTT is considered to be a 6-cutter.
Output
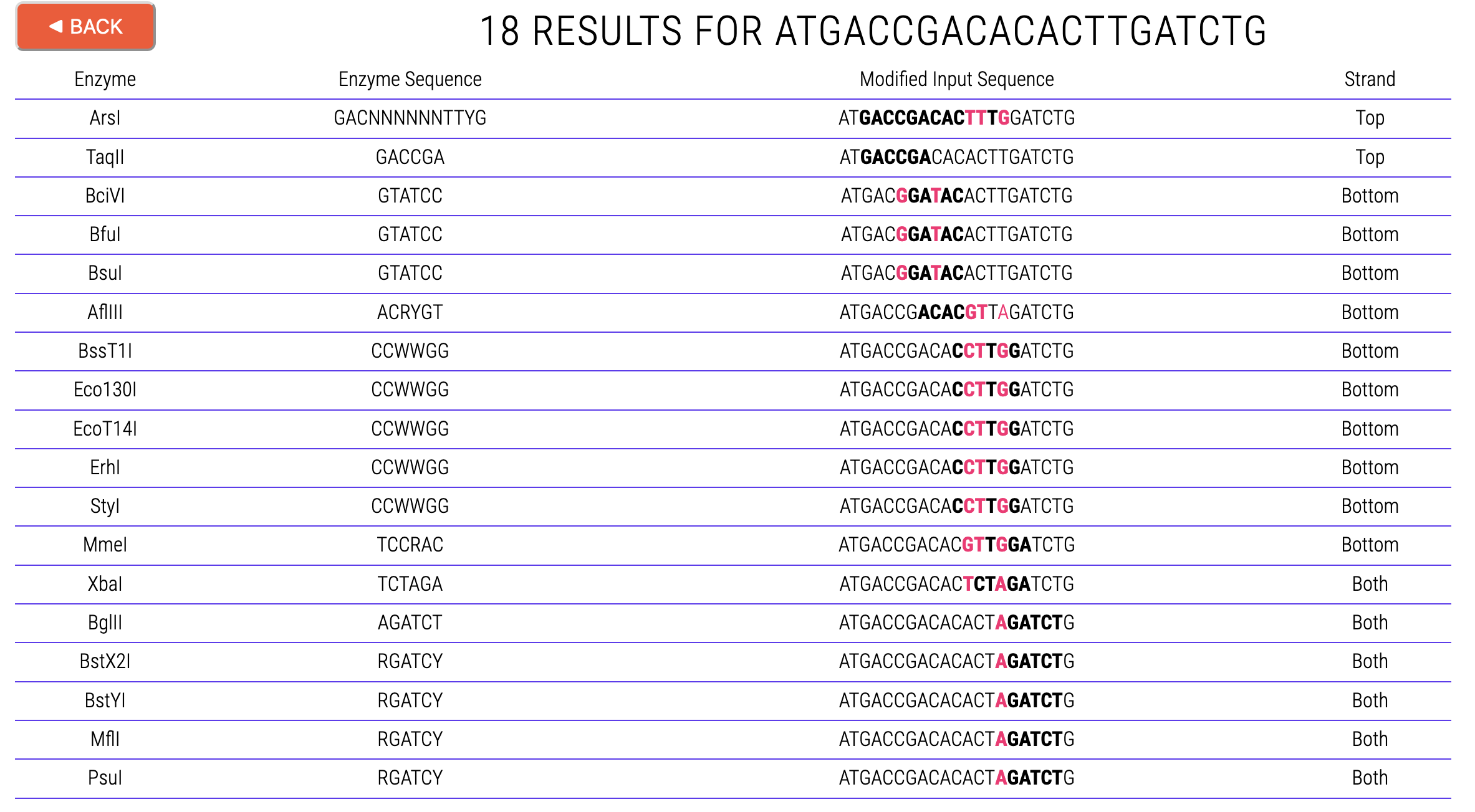
An example output for a 21 bp DNA sequence (5’–ATGACCGACACACTTGATCTG–3’) encoding a 7 aa peptide (MTDTLDL) is shown above. In this case, 6-cutters and up were chosen.
- Nucleotides in BOLD indicate the restriction enzyme site. In some cases (e.g., ArsI) these may include non-specific “spacer” residues (N).
- Nucleotides in RED indicate residues that were changed to create the new restriction site while preserving the coding sequence. In some cases (e.g., AflIII) changes may be made just outside the enzyme recognition site to retain the original peptide sequence.
- Both the enzyme name and the consensus recognition sequence are shown.
- For “bottom strand” (non-reverse-palindromic) enzymes, the recognition site will be the reverse complement of the displayed top strand sequence.
- Isoschizomers, different enzymes that have the same recognition sequence, are displayed on different lines. In some cases, only one may be commercially available.
- In the case of enzymes containing non-specific nucleotides (e.g., R,Y) as part of their recognition sequence, only one example sequence is provided. However, know that there may be additional iterations that could potentially work.
- A=Adenine G=Guanine C=Cytosine T=Thymine
N=Any nucleotide
R=A or G
Y=C or T
W=A or T
S=G or C
M=A or C
K=G or T
B=G or C or T
H=A or C or T
D=A or G or T
V=A or G or C
What CRISPRcruncher Does Not Do
- CRISPRcruncher will not suggest a PAM site. Other tools are available to “help” with this, and the optimal PAM/cut site will likely depend on multiple variables. It’s always something of a gamble.
- CRISPRcruncher will not suggest the best location for the new restriction site or the best configuration for the various changes you may wish to make as part of your repair template design. As for the PAM site, this part of the process can depend on multiple variables and you will want to consult the latest CRISPR literature.
- CRISPRcruncher will not provide specific suggestions for ablating a site already present in the starting sequence. This is easy enough to do manually.
- CRISPRcruncher does not take into account possible codon bias (codon usage frequency). This issue varies from organism to organism and is one of the multiple variables that may need to be factored into your design.
- CRISPRcruncher will not write your prelim, paper, thesis, grant, or do your dishes for you, despite claims to the contrary. We have no idea where those rumors even got started.
Citing Our Paper
Please cite our paper!
Fay SF, Fay DS, Chhatre VE. (2021) CRISPRcruncher: A tool for engineering restriction sites into coding regions. MicroPubl Biol. 2021 Jan 18;2021:10.17912/micropub.biology.000343. doi: 10.17912/micropub.biology.000343. PMID: 33490886
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7816087/